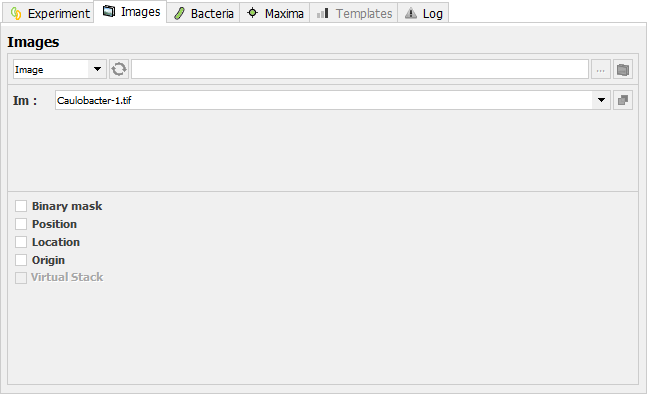
This panel contains the settings relative to the images to be analyzed. Press ![]() to update the list of the opened images in dropdown menus. Press
to update the list of the opened images in dropdown menus. Press ![]() to locate the directory containing the Image(s) to be analyzed. Press
to locate the directory containing the Image(s) to be analyzed. Press ![]() to load the specified images into a hyperstack. Note that in this case, MicrobeJ will turn automatically into the Image input mode.
to load the specified images into a hyperstack. Note that in this case, MicrobeJ will turn automatically into the Image input mode.
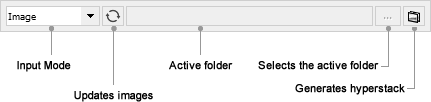
Input Mode: Allows any of the five different image input modes to be selected: Current, Image, Channel(s), Folder, Folder Channel. Five modes are available to select single images, one-dimensional stacks or multi-dimensional stacks (i.e. hyperstacks) or load them from disk.
Current: Selects the active opened image in ImageJ.
Image: Selects one opened image, stack or hyperstack to be analyzed using the Im dropdown menu. Note that both uni-dimensional and multi-dimensional stacks are listed in the dropdown menu.
Channel(s): Selects 1-4 opened single images or one-dimensional stacks to be analyzed. Select the associated channel using the Ch.1-Ch.4 dropdown menus. Select *none* to skip a channel. Note that only opened single images or one-dimensional stacks are listed in the dropdown menus.
Folder: Opens a series of images, stacks or hyperstacks from a chosen folder as a hyperstack. Enter any sequence of characters the filename must contain to be loaded using the Im field. Leave the field empty to load all the images located into the specified directory. Images can be of any format supported by ImageJ (please refer to the ImageJ user’s manual section 7 Image Types and Formats). Non-image files are ignored. Images must have the same dimensions (width x height x channel) but can have different number of frame/slices. Note that stacks will be opened in numeric file name order (e.g., ‘name1.tif’, ‘name2.tif’, ‘name10.tif’). Press ![]() to load the specified images into a hyperstack. Note that it is not necessary to load the specified images to run the experiment.
to load the specified images into a hyperstack. Note that it is not necessary to load the specified images to run the experiment.
Folder Channel: Opens a series of single images or one-dimensional stacks from a chosen folder as a stack or a hyperstack. Enter any sequence of characters the filename must contain to be associated with a specific channel using the Ch.1-Ch.4 field. Images can be of any format supported by ImageJ. Non-image files are ignored. Images must have the same dimensions (width x height) but can have different number of frame/slices. Note that stacks will be opened in numeric file name order (e.g., ‘name1.tif’, ‘name2.tif’, ‘name10.tif’). Press ![]() to load the specified images into a hyperstack. Note that it is not necessary to load the specified images to run the experiment.
to load the specified images into a hyperstack. Note that it is not necessary to load the specified images to run the experiment.
Options:
Binary Mask: If checked, a specified binary image or stack can be used instead of automatic thresholding. Note that only opened binary images/stack are listed in the dropdown menu.
Position: If checked, analysis will be performed only on the specified range of Slices (z) and/or Frames (t).
Location: If checked, analysis will be performed only on the specified region of interest.
Origin: If checked, select the location of the origin in the Image. Default values are 0.
Virtual Stack: If checked, loads the specified images as a Virtual Stack. Virtual stacks are disk resident and are the only way to load image sequences that do not fit in RAM.
Created with the Personal Edition of HelpNDoc: Full-featured EBook editor